Advanced Machine Learning - Regression Pipelines in Scikit-learn
In this third part of our series, we’ll explore more sophisticated machine learning techniques using Scikit-learn. While Parts 1 and 2 focused on classification, we’ll now tackle regression problems and learn how to build complex preprocessing pipelines. We’ll use the California Housing dataset to demonstrate these concepts.
The complete code for this tutorial can be found in the 03_scikit_advanced.py script.
Note: The purpose of this post is to highlight the flexibility and capabilities of scikit-learn’s advanced features. Therefore, this tutorial focuses on introducing you to those advanced routines rather than creating the optimal regression model.
Why Advanced Preprocessing?
Real-world data rarely comes in a clean, ready-to-use format. Data scientists often spend more time preparing data than training models. Common preprocessing steps include:
- Missing value imputation: Filling missing data points
- Feature encoding: Converting categorical variables to numerical format
- Feature scaling: Normalizing features to comparable ranges
- Feature selection: Identifying most relevant variables
- Feature engineering: Creating new features from existing ones
Scikit-learn provides powerful tools to handle these challenges systematically. Let’s see how to combine them effectively into a preprocessing pipeline that can handle all these issues automatically.
As always, first, let’s import the scientific Python packages we need.
# Standard scientific Python imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
1. Load Dataset
The California Housing dataset contains information about houses in California districts. It’s a perfect dataset for demonstrating advanced preprocessing because it includes:
- Both numerical and categorical features
- Missing values that need handling
- Features on different scales
- Complex relationships between variables
The dataset itself contains information about the houses, including features like total area, lot shape, neighborhood information, overall quality, year built, etc. And the target feature that we would like to predict is the SalePrice.
Let’s load the data and take a look:
# Load dataset
from sklearn import datasets
housing = datasets.fetch_openml(name='house_prices')
# Extract feature matrix X and target vector y
X = housing['data'].drop(columns='Id')
y = housing['target']
print(f"Dimension of X: {X.shape}\nDimension of y: {y.shape}")
Dimension of X: (1460, 79)
Dimension of y: (1460,)
The house price dataset contains:
- 1,460 samples: Each representing a different house sale
-
79 features: A mix of numerical and categorical characteristics including:
- Property specifications (size, rooms, year built)
- Location details (neighborhood, zoning)
- Quality ratings (overall condition, materials)
- Target values: Continuous house sale prices in dollars
As you can see, we have 1460 samples (houses), each containing 79 features (i.e. characteristics). Let’s examine the first few entries to better understand our data:
# Show first few entries and columns of the dataset
X.iloc[:5, :5]
If we look closer at the feature matrix X, we can see that of those 79 features, 36 are of type float and 43
are of type ‘object’ (i.e. categorical features), and that some entries are missing. Plus, the target feature
SalePrice has a right skewed value distribution.
Therefore, if possible, our pipeline should be able to handle all of this peculiarities. Even better, let’s try to setup a pipeline that helps us to find the optimal way how to preprocess this dataset.
2. Feature Analysis
Before building our pipeline, let’s understand what we’re working with:
# Quick overview of feature types
print("Feature types:")
print(X.dtypes.value_counts())
# Check for missing values
missing_values = X.isnull().sum()
print("\nFeatures with missing values:")
print(missing_values[missing_values > 0].sort_values(ascending=False))
Feature types:
object 43
int64 33
float64 3
Name: count, dtype: int64
Features with missing values:
PoolQC 1453
MiscFeature 1406
Alley 1369
Fence 1179
FireplaceQu 690
LotFrontage 259
GarageType 81
GarageYrBlt 81
GarageFinish 81
GarageQual 81
GarageCond 81
BsmtExposure 38
BsmtFinType2 38
BsmtFinType1 37
BsmtCond 37
BsmtQual 37
MasVnrArea 8
MasVnrType 8
Electrical 1
dtype: int64
And visualizing the target variable distribution:
# Analyze target variable distribution
plt.figure(figsize=(8, 4))
sns.histplot(y, bins=50)
plt.title("Distribution of House Prices")
plt.xlabel("Price")
plt.show()
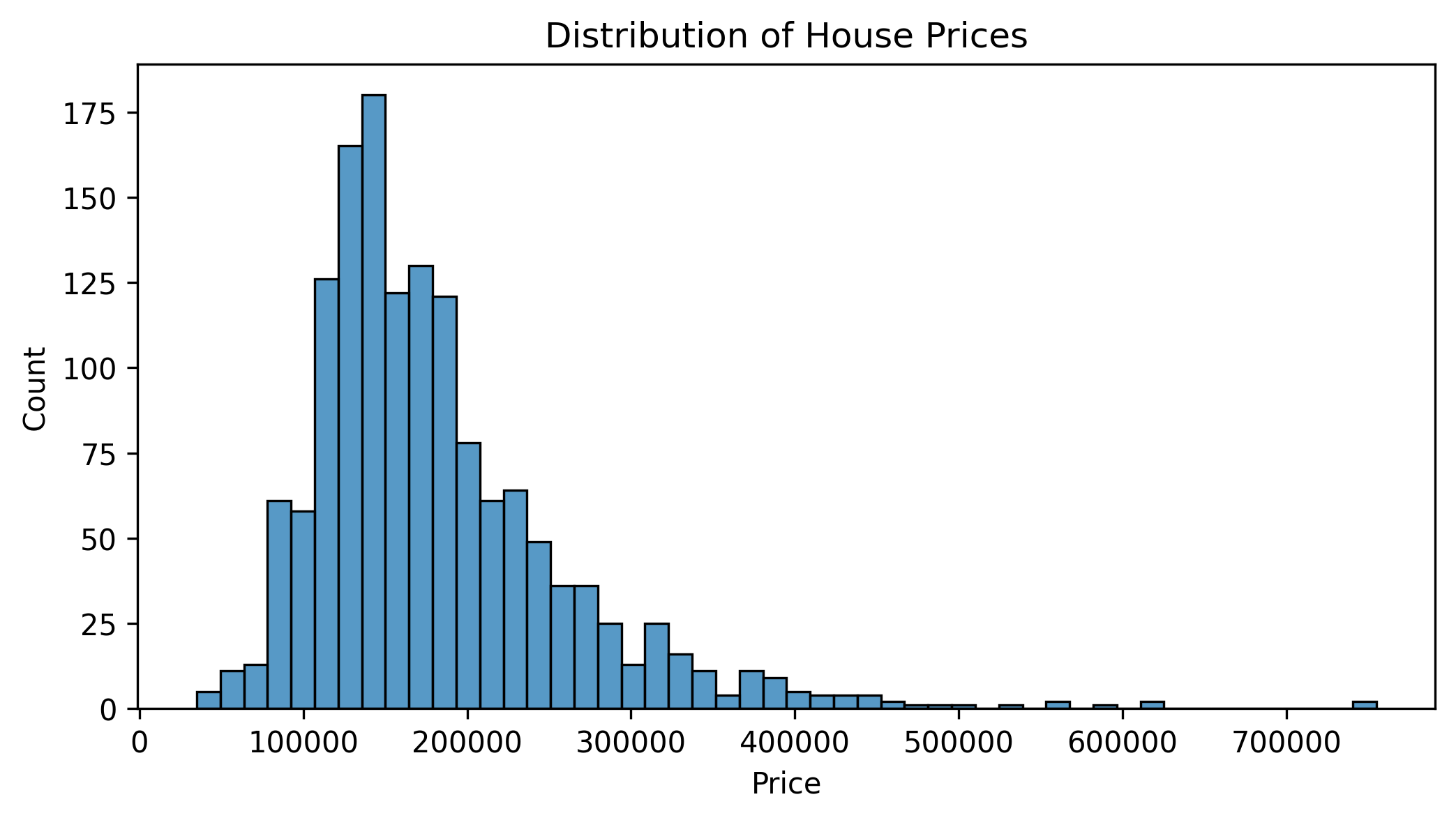
This analysis reveals several important preprocessing needs:
- We have both numerical (float) and categorical (object) features
- Several features have missing values
- Our target variable (house prices) shows right skew
- Features are on very different scales (e.g., year vs. price)
These insights will guide our pipeline design.
3. Split data into train and test set
As always, let’s first go ahead and split the dataset into train and test set.
from sklearn.model_selection import train_test_split
X_tr, X_te, y_tr, y_te = train_test_split(X, y, test_size=0.2)
4. Building the Pipeline
One of Scikit-learn’s most powerful features is its Pipeline API. We’ll create a pipeline that:
- Handles missing values differently for numerical and categorical features
- Applies appropriate scaling to numerical features
- Properly encodes categorical features
- Optionally reduces dimensionality
- Fits our chosen regression model
So let’s setup a pipeline that performs these different pre-processing routines: Transformation of categorical data to numerical data, data imputer for missing values, data scaling, potential dimensionality reduction, etc.
4.1. Handling categorical data
First, let’s create a small pipeline that takes categorical data, fills missing values with 'missing' and
than applies one-hot encoding on these categorical features.
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import OneHotEncoder
# Define preprocessing pipeline for categorical features
categorical_preprocessor = Pipeline(
[
# Fill missing values with 'missing' string
('imputer_cat', SimpleImputer(fill_value='missing', strategy='constant')),
# Convert categorical strings to one-hot encoded vectors
# handle_unknown='ignore' prevents errors with new categories at prediction time
('onehot', OneHotEncoder(handle_unknown='ignore')),
]
)
4.2. Handling numerical data
To handle numerical data we will use a slightly more advanced processing pipeline (to showcase some scikit-learn feature, not because it’s the best thing to do). So let’s first fill missing values with e.g. the mean of the feature, potentially apply a polynomial expansion to module non-linear relationships, apply a scaler and then potentially apply dimensionality reduction via PCA and/or by selecting only the “most relevant” features.
from sklearn.decomposition import PCA
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression, mutual_info_regression
from sklearn.pipeline import FeatureUnion
# Create feature reduction pipeline combining PCA and feature selection
dim_reduction = FeatureUnion(
[
('pca', PCA()), # Dimensionality reduction using principal component analysis
('feat_selecter', SelectKBest()), # Select top K features based on statistical tests
]
)
from sklearn.preprocessing import (PolynomialFeatures, StandardScaler,
RobustScaler, PowerTransformer)
# Package all relevant preprocessing routines for numerical data into one pipeline
numeric_preprocessor = Pipeline(
[
# Handle missing values in numerical features
('imputer_numeric', SimpleImputer(
missing_values=np.nan, # Identify NaN values
strategy='mean')), # Replace with column mean
('polytrans', PolynomialFeatures()), # Create interaction terms between features
('scaler', StandardScaler()), # Normalize features to zero mean and unit variance
('dim_reduction', dim_reduction), # Apply dimensionality reduction
]
)
4.3. Combining preprocessing pipelines
Now that we have a preprocessing pipeline for the categorical and numerical features, let’s combine them into one preprocessing pipeline.
from sklearn.compose import ColumnTransformer
preprocessor = ColumnTransformer(
[
('numerical', numeric_preprocessor, X.select_dtypes('number').columns),
('categorical', categorical_preprocessor, X.select_dtypes(exclude='number').columns),
],
remainder='passthrough',
)
4.4. Add regression model
After the data is preprocessed we want to hand it over to a regression estimator. For this purpose, let’s chose a ridge regression.
from sklearn.linear_model import Ridge
ridge = Ridge()
pipe = Pipeline(steps=[
('preprocessor', preprocessor),
('ridge', Ridge())
])
As such, the pipeline would be finished. But because we know that our target feature SalePrice is right
skewed, we should ideally apply a log-transformation before fitting the model. Instead of doing this
transformation manually (and reverting it at the end), we can also use scikit-learn’s
TransformedTargetRegressor to do that on the fly.
from sklearn.compose import TransformedTargetRegressor
regressor = TransformedTargetRegressor(
regressor=pipe, func=np.log1p, inverse_func=np.expm1
)
5. Parameter grid
Before training our model, we should also define a parameter grid that allows us to fine-tune the processing and model parameters. Given our complex routine, we actually have a lot of parameter that we can play around with.
from sklearn.model_selection import ParameterGrid
# Shorten key identifier by separating common prefix
prefix = 'regressor__preprocessor__'
# Create parametergrid
param_grid = {
# Explore imputers
f'{prefix}numerical__imputer_numeric__add_indicator': [True, False],
f'{prefix}numerical__imputer_numeric__strategy': [
'mean', 'median', 'most_frequent', 'constant'],
f'{prefix}categorical__imputer_cat__add_indicator': [True, False],
f'{prefix}categorical__imputer_cat__strategy': ['most_frequent', 'constant'],
# Explore numerical preprocessors
f'{prefix}numerical__polytrans__degree': [1, 2],
f'{prefix}numerical__polytrans__interaction_only': [False, True],
f'{prefix}numerical__dim_reduction__pca': ['drop', PCA(0.9), PCA(0.99)],
f'{prefix}numerical__dim_reduction__feat_selecter__k': [5, 25, 100, 'all'],
f'{prefix}numerical__dim_reduction__feat_selecter__score_func': [
f_regression, mutual_info_regression],
# Explore scalers
f'{prefix}numerical__scaler': [StandardScaler(), RobustScaler(), PowerTransformer()],
# Explore regressor
'regressor__ridge__alpha': np.logspace(-5, 5, 11),
}
print(len(ParameterGrid(param_grid)))
101376
As you can see, we have more than 100’000 different parameter combinations that we could explore. So using a
GridSearchCV routine and checking all of them individually would take way too much time. Luckily,
scikit-learn also provides a RandomizedSearchCV routine, with which you can randomly explore a few parameter
grid combinations.
Furthermore, both GridSearchCV and RandomizedSearchCV routines also allow you to change the performance
metric with which the model performs is scored. So let’s take 'neg_mean_absolute_percentage_error' (for more
see here).
from sklearn.model_selection import RandomizedSearchCV
random_search = RandomizedSearchCV(
regressor,
param_grid,
n_iter=250,
refit=True,
cv=2,
return_train_score=True,
n_jobs=-1,
verbose=1,
scoring='neg_mean_absolute_percentage_error',
)
6. Train model
Everything is ready, so let’s go ahead and train the model.
res = random_search.fit(X_tr, y_tr)
Fitting 2 folds for each of 250 candidates, totalling 500 fits
7. Performance investigation after RandomizedSearchCV
Once the model has explored a fixed number of grid points, we can go ahead and look at their performance. The easiest is to just put everything into a pandas DataFrame and sort the entries by the best test score.
# Create dataframe with results
df_res = pd.DataFrame(res.cv_results_)
# Remove columns that are not relevant for the analysis
df_res = df_res.iloc[:, ~df_res.columns.str.contains('time|split[0-9]*|rank|params')]
# Rename columns to make them more readable
new_columns = [c.split('param_regressor__')[1] if 'param_regressor' in c else c for c in df_res.columns]
new_columns = [c.split('preprocessor__')[1] if 'preprocessor__' in c else c for c in new_columns]
df_res.columns = new_columns
df_res = df_res.sort_values('mean_test_score', ascending=False)
print("\nTop 10 parameter combinations:")
print(df_res.head(10))
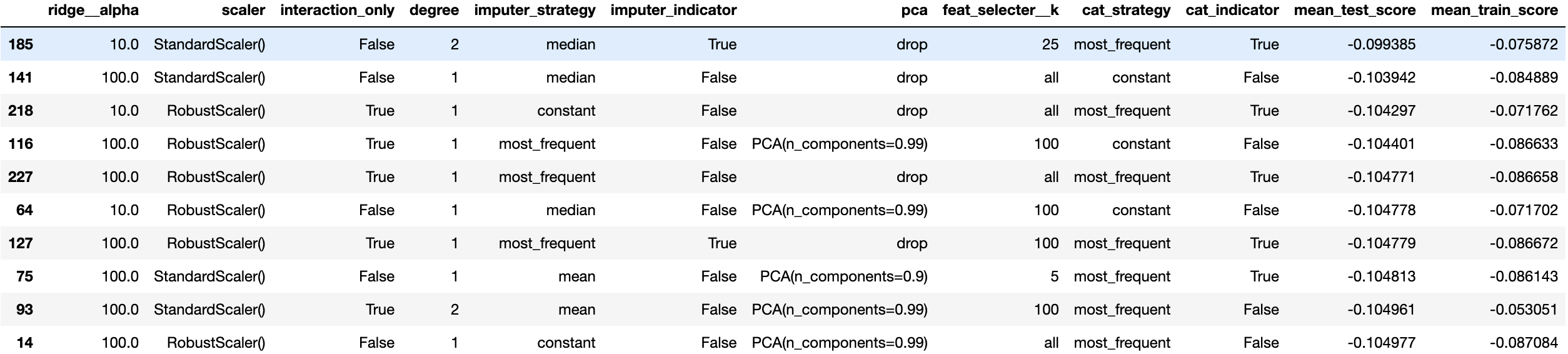
If you explore this table a bit you can better judge which parameter variations in your grid search are actually useful and which ones aren’t. In this example we will not focus on this and directly continue with computing the model performance on the training and test set.
# Evaluate model performance on training and test set
score_tr = -random_search.score(X_tr, y_tr)
score_te = -random_search.score(X_te, y_te)
print(
f"Prediction accuracy on train data: {score_tr*100:.2f}%\n\
Prediction accuracy on test data: {score_te*100:.2f}%"
)
Prediction accuracy on train data: 7.08%
Prediction accuracy on test data: 9.09%
Let’s interpret these regression metrics in practical terms:
-
Train Error: On average, predictions deviate by about 7-8% from true house prices
- For a $300,000 house, this means predictions are typically within ±$21,000-24,000
-
Test Error: Slightly higher error on unseen data
- For a $300,000 house, predictions are typically within ±$24,000-27,000
- Error Difference: Small gap indicates good generalization
- Context: For house price prediction, ~8-9% error is relatively good considering market volatility
Great, the score seems reasonably good! But now that we know better which preprocessing routine seems to be the
best (thanks to RandomizedSearchCV), let’s go ahead and further fine-tune the ridge model.
8. Fine tune best preprocessing pipeline
To further fine tune the best preprocessing pipeline, we can just load the ‘best estimator’ from the
RandomizedSearchCV exploration and specify a new parameter grid that we want to explore - this time with the
GridSearchCV routine (so that we look at all grid points).
# Select best estimator
best_estimator = random_search.best_estimator_
# Specify new parameter grid to explore
param_grid = {'regressor__ridge__alpha': np.logspace(-5, 5, 51)}
To showcase one more additional thing, let’s go ahead and use a nested cross-validation routine to improve the generalization power of our model. In other words, in contrast to the previous approach where we separated the test from the train set only once, we will now also apply a cross validation approach on this split as well. Together with the cross validation in the grid search, we therefore use cross validation twice, hence the name “nested”.
# Establish the two cross validations
from sklearn.model_selection import KFold
inner_cv = KFold(n_splits=3, shuffle=True)
outer_cv = KFold(n_splits=3, shuffle=True)
And now, let’s combine all of this with and run the model.
from sklearn.model_selection import GridSearchCV, cross_validate
# Create grid search object with parameter grid and inner cross validation
grid_search = GridSearchCV(
best_estimator,
param_grid,
refit=True,
cv=inner_cv,
n_jobs=-1,
scoring='neg_mean_absolute_percentage_error',
)
# Train model with outer cross validation (and return estimators for post-model investigation)
cv_results = cross_validate(
grid_search,
X=X,
y=y,
cv=outer_cv,
n_jobs=1,
return_estimator=True,
return_train_score=True,
)
Once the model has finished training, we can extract the different scores from the most outer loop and print their average score, as well as the standard deviation over the folds. Plus, the same thing can also be done for the most optimal ridge model parameter ‘alpha’. These information can give us some insights about the model generalization.
df_nested = pd.DataFrame(cv_results)
cv_train_scores = -df_nested['train_score']
cv_test_scores = -df_nested['test_score']
cv_alphas = [c.best_params_['regressor__ridge__alpha'] for c in df_nested['estimator']]
print(
"Generalization score with hyperparameters tuning:\n"
f" Train Score: {cv_train_scores.mean()*100:.1f}% +/- {cv_train_scores.std()*100:.1f}%\n"
f" Test Score: {cv_test_scores.mean()*100:.1f}% +/- {cv_test_scores.std()*100:.1f}%\n"
f" Optimal Alpha: {np.mean(cv_alphas):.1f} +/- {np.std(cv_alphas):.1f}\n"
)
Generalization score with hyperparameters tuning:
Train Score: 7.6% +/- 0.9%
Test Score: 9.0% +/- 0.2%
Optimal Alpha: 29.6 +/- 23.8
9. Feature importance investigation with permutation testing
Some model provide some insights about feature importance (i.e. which features the model uses most for the prediction). However, this is sometimes prone to multiple issues. A better approach is to use a permutation approach. This approach performs the same model fitting (in this case based on the best model with the best hyper parameters) but during each iteration randomly shuffles a given feature and investigates how this perturbates the final score.
# Select the best estimator with the best hyper parameter
final_estimator = grid_search.set_params(
estimator__regressor__ridge__alpha=np.mean(cv_alphas)
)
# Fit this estimator to the initial training set
_ = final_estimator.fit(X_tr, y_tr)
Now that the model is ready and trained, we can go ahead and perform the feature importance investigation via
permutation testing. To showcase one additional feature, let’s actually perform this routine twice, once while
focusing on the r2 of the model, and once while focusing on the neg_mean_absolute_percentage_error.
from sklearn.inspection import permutation_importance
scoring = ['r2', 'neg_mean_absolute_percentage_error']
result = permutation_importance(
final_estimator,
X_te,
y_te,
n_repeats=50,
random_state=0,
n_jobs=-1,
scoring=scoring,
)
Once everything is computed, we can go ahead and plot the feature importance for each feature, separated by the two different scoring metrics.
fig, axs = plt.subplots(1, 2, figsize=(16, 16))
for i, s in enumerate(scoring):
sorted_idx = result[s].importances_mean.argsort()
axs[i].boxplot(
result[s].importances[sorted_idx].T, vert=False, labels=X_te.columns[sorted_idx]
)
axs[i].set_title("Permutation Importances (test set) | %s" % s)
fig.tight_layout()
plt.show()
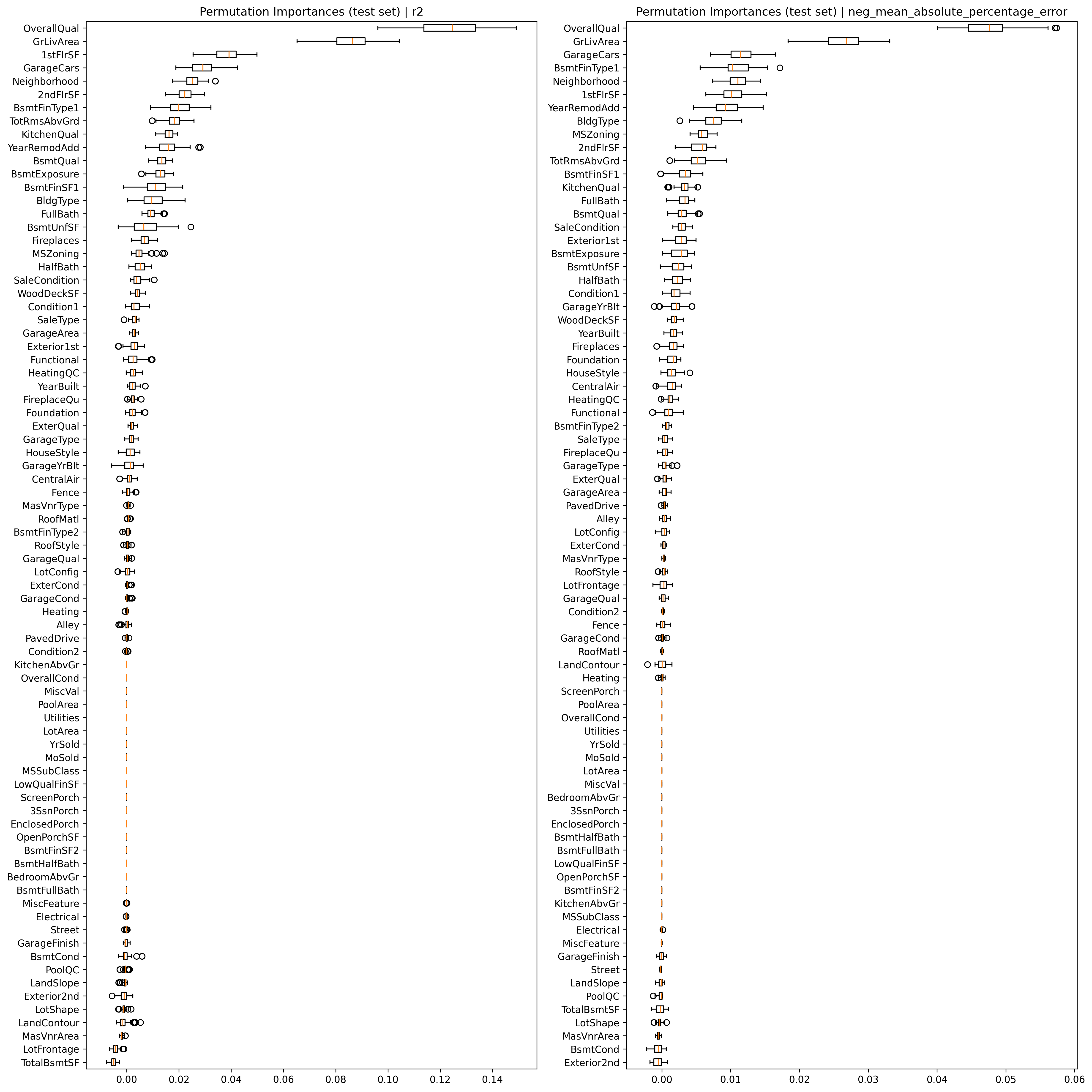
Summary and Next Steps
In this tutorial, we’ve covered advanced scikit-learn concepts:
- Building complex preprocessing pipelines
- Handling mixed data types
- Feature selection and engineering automatically
- Implementing grid search with cross-validation
- Model comparison and evaluation
- Analyzing feature importance
Key takeaways:
- Preprocessing pipelines make complex workflows manageable
- Grid search helps find optimal parameters systematically
- Feature selection can improve model performance
- Understanding feature importance aids model interpretation
- Cross-validation provides robust performance estimates
In Part 4, we’ll explore advanced neural network architectures with TensorFlow, building on both the neural network concepts from Part 2 and the preprocessing techniques we’ve learned here.
← Previous: Deep Learning Fundamentals or Next: Advanced Deep Learning →