JoinNode, synchronize and itersource¶
JoinNode has the opposite effect of iterables. Where iterables split up the execution workflow into many different branches, a JoinNode merges them back into on node. A JoinNode generalizes MapNode to operate in conjunction with an upstream iterable node to reassemble downstream results, e.g.:
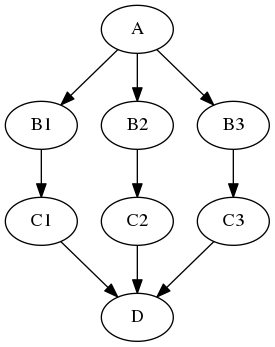
Simple example¶
Let's consider the very simple example depicted at the top of this page:
from nipype import Node, JoinNode, Workflow
# Specify fake input node A
a = Node(interface=A(), name="a")
# Iterate over fake node B's input 'in_file?
b = Node(interface=B(), name="b")
b.iterables = ('in_file', [file1, file2])
# Pass results on to fake node C
c = Node(interface=C(), name="c")
# Join forked execution workflow in fake node D
d = JoinNode(interface=D(),
joinsource="b",
joinfield="in_files",
name="d")
# Put everything into a workflow as usual
workflow = Workflow(name="workflow")
workflow.connect([(a, b, [('subject', 'subject')]),
(b, c, [('out_file', 'in_file')])
(c, d, [('out_file', 'in_files')])
])
As you can see, setting up a JoinNode is rather simple. The only difference to a normal Node is the joinsource and the joinfield. joinsource specifies from which node the information to join is coming and the joinfield specifies the input field of the JoinNode where the information to join will be entering the node.
This example assumes that interface A has one output subject, interface B has two inputs subject and in_file and one output out_file, interface C has one input in_file and one output out_file, and interface D has one list input in_files. The images variable is a list of three input image file names.
As with iterables and the MapNode iterfield, the joinfield can be a list of fields. Thus, the declaration in the previous example is equivalent to the following:
d = JoinNode(interface=D(),
joinsource="b",
joinfield=["in_files"],
name="d")
The joinfield defaults to all of the JoinNode input fields, so the declaration is also equivalent to the following:
d = JoinNode(interface=D(),
joinsource="b",
name="d")
In this example, the node C out_file outputs are collected into the JoinNode D in_files input list. The in_files order is the same as the upstream B node iterables order.
The JoinNode input can be filtered for unique values by specifying the unique flag, e.g.:
d = JoinNode(interface=D(),
joinsource="b",
unique=True,
name="d")
synchronize¶
The Node iterables parameter can be be a single field or a list of fields. If it is a list, then execution is performed over all permutations of the list items. For example:
b.iterables = [("m", [1, 2]), ("n", [3, 4])]
results in the execution graph:
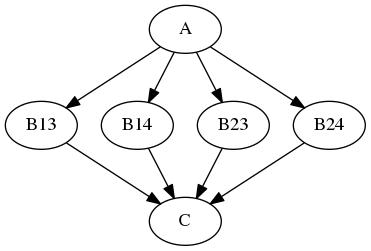
where B13 has inputs m = 1, n = 3, B14 has inputs m = 1, n = 4, etc.
The synchronize parameter synchronizes the iterables lists, e.g.:
b.iterables = [("m", [1, 2]), ("n", [3, 4])]
b.synchronize = True
results in the execution graph:
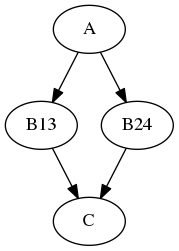
where the iterable inputs are selected in lock-step by index, i.e.:
(*m*, *n*) = (1, 3) and (2, 4)
for B13 and B24, resp.
itersource¶
The itersource feature allows you to expand a downstream iterable based on a mapping of an upstream iterable. For example:
a = Node(interface=A(), name="a")
b = Node(interface=B(), name="b")
b.iterables = ("m", [1, 2])
c = Node(interface=C(), name="c")
d = Node(interface=D(), name="d")
d.itersource = ("b", "m")
d.iterables = [("n", {1:[3,4], 2:[5,6]})]
my_workflow = Workflow(name="my_workflow")
my_workflow.connect([(a,b,[('out_file','in_file')]),
(b,c,[('out_file','in_file')])
(c,d,[('out_file','in_file')])
])
results in the execution graph:
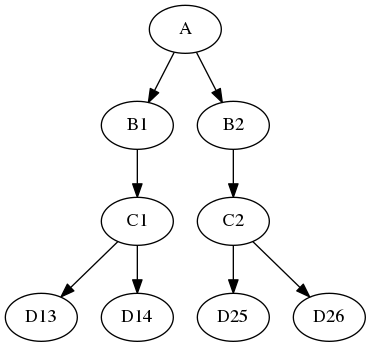
In this example, all interfaces have input in_file and output out_file. In addition, interface B has input m and interface D has input n. A Python dictionary associates the B node input value with the downstream D node n iterable values.
This example can be extended with a summary JoinNode:
e = JoinNode(interface=E(), joinsource="d",
joinfield="in_files", name="e")
my_workflow.connect(d, 'out_file',
e, 'in_files')
resulting in the graph:
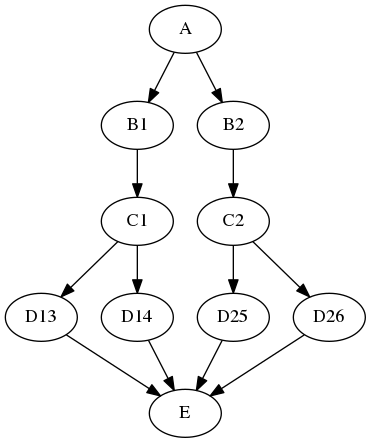
The combination of iterables, MapNode, JoinNode, synchronize and itersource enables the creation of arbitrarily complex workflow graphs. The astute workflow builder will recognize that this flexibility is both a blessing and a curse. These advanced features are handy additions to the Nipype toolkit when used judiciously.
More realistic JoinNode example¶
Let's consider another example where we have one node that iterates over 3 different numbers and generates random numbers. Another node joins those three different numbers (each coming from a separate branch of the workflow) into one list. To make the whole thing a bit more realistic, the second node will use the Function interface to do something with those numbers, before we spit them out again.
from nipype import JoinNode, Node, Workflow
from nipype.interfaces.utility import Function, IdentityInterface
def get_data_from_id(id):
"""Generate a random number based on id"""
import numpy as np
return id + np.random.rand()
def merge_and_scale_data(data2):
"""Scale the input list by 1000"""
import numpy as np
return (np.array(data2) * 1000).tolist()
node1 = Node(Function(input_names=['id'],
output_names=['data1'],
function=get_data_from_id),
name='get_data')
node1.iterables = ('id', [1, 2, 3])
node2 = JoinNode(Function(input_names=['data2'],
output_names=['data_scaled'],
function=merge_and_scale_data),
name='scale_data',
joinsource=node1,
joinfield=['data2'])
wf = Workflow(name='testjoin')
wf.connect(node1, 'data1', node2, 'data2')
eg = wf.run()
wf.write_graph(graph2use='exec')
from IPython.display import Image
Image(filename='graph_detailed.png')
Now, let's look at the input and output of the joinnode:
res = [node for node in eg.nodes() if 'scale_data' in node.name][0].result
res.outputs
res.inputs
Extending to multiple nodes¶
We extend the workflow by using three nodes. Note that even this workflow, the joinsource corresponds to the node containing iterables and the joinfield corresponds to the input port of the JoinNode that aggregates the iterable branches. As before the graph below shows how the execution process is set up.
def get_data_from_id(id):
import numpy as np
return id + np.random.rand()
def scale_data(data2):
import numpy as np
return data2
def replicate(data3, nreps=2):
return data3 * nreps
node1 = Node(Function(input_names=['id'],
output_names=['data1'],
function=get_data_from_id),
name='get_data')
node1.iterables = ('id', [1, 2, 3])
node2 = Node(Function(input_names=['data2'],
output_names=['data_scaled'],
function=scale_data),
name='scale_data')
node3 = JoinNode(Function(input_names=['data3'],
output_names=['data_repeated'],
function=replicate),
name='replicate_data',
joinsource=node1,
joinfield=['data3'])
wf = Workflow(name='testjoin')
wf.connect(node1, 'data1', node2, 'data2')
wf.connect(node2, 'data_scaled', node3, 'data3')
eg = wf.run()
wf.write_graph(graph2use='exec')
Image(filename='graph_detailed.png')
Exercise 1¶
You have list of DOB of the subjects in a few various format : ["10 February 1984", "March 5 1990", "April 2 1782", "June 6, 1988", "12 May 1992"], and you want to sort the list.
You can use Node with iterables to extract day, month and year, and use datetime.datetime to unify the format that can be compared, and JoinNode to sort the list.
# write your solution here
# the list of all DOB
dob_subjects = ["10 February 1984", "March 5 1990", "April 2 1782", "June 6, 1988", "12 May 1992"]
# let's start from creating Node with iterable to split all strings from the list
from nipype import Node, JoinNode, Function, Workflow
def split_dob(dob_string):
return dob_string.split()
split_node = Node(Function(input_names=["dob_string"],
output_names=["split_list"],
function=split_dob),
name="splitting")
#split_node.inputs.dob_string = "10 February 1984"
split_node.iterables = ("dob_string", dob_subjects)
# and now let's work on the date format more, independently for every element
# sometimes the second element has an extra "," that we should remove
def remove_comma(str_list):
str_list[1] = str_list[1].replace(",", "")
return str_list
cleaning_node = Node(Function(input_names=["str_list"],
output_names=["str_list_clean"],
function=remove_comma),
name="cleaning")
# now we can extract year, month, day from our list and create ``datetime.datetim`` object
def datetime_format(date_list):
import datetime
# year is always the last
year = int(date_list[2])
#day and month can be in the first or second position
# we can use datetime.datetime.strptime to convert name of the month to integer
try:
day = int(date_list[0])
month = datetime.datetime.strptime(date_list[1], "%B").month
except(ValueError):
day = int(date_list[1])
month = datetime.datetime.strptime(date_list[0], "%B").month
# and create datetime.datetime format
return datetime.datetime(year, month, day)
datetime_node = Node(Function(input_names=["date_list"],
output_names=["datetime"],
function=datetime_format),
name="datetime")
# now we are ready to create JoinNode and sort the list of DOB
def sorting_dob(datetime_list):
datetime_list.sort()
return datetime_list
sorting_node = JoinNode(Function(input_names=["datetime_list"],
output_names=["dob_sorted"],
function=sorting_dob),
joinsource=split_node, # this is the node that used iterables for x
joinfield=['datetime_list'],
name="sorting")
# and we're ready to create workflow
ex1_wf = Workflow(name="sorting_dob")
ex1_wf.connect(split_node, "split_list", cleaning_node, "str_list")
ex1_wf.connect(cleaning_node, "str_list_clean", datetime_node, "date_list")
ex1_wf.connect(datetime_node, "datetime", sorting_node, "datetime_list")
# you can check the graph
from IPython.display import Image
ex1_wf.write_graph(graph2use='exec')
Image(filename='graph_detailed.png')
# and run the workflow
ex1_res = ex1_wf.run()
# you can check list of all nodes
ex1_res.nodes()
# and check the results from sorting_dob.sorting
list(ex1_res.nodes())[0].result.outputs