BRAIN IMAGING
DATA STRUCTURE
The dataset for this tutorial is structured according to the Brain Imaging Data Structure (BIDS). BIDS is a simple and intuitive way to organize and describe your neuroimaging and behavioral data. Neuroimaging experiments result in complicated data that can be arranged in many different ways. So far there is no consensus on how to organize and share data obtained in neuroimaging experiments. BIDS tackles this problem by suggesting a new standard for the arrangement of neuroimaging datasets.
The idea of BIDS is that the file and folder names follow a strict set of rules:
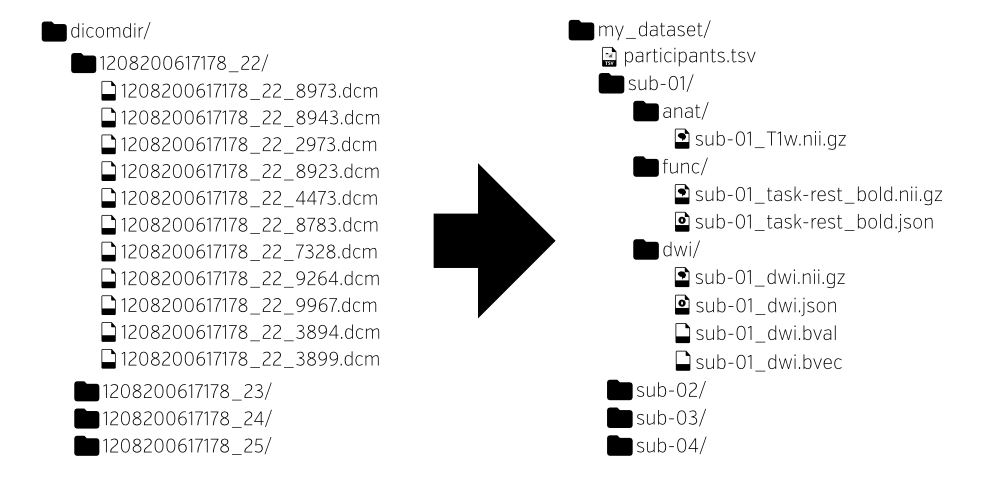
Using the same structure for all of your studies will allow you to easily reuse all of your scripts between studies. But additionally, it also has the advantage that sharing code with and using scripts from other researchers will be much easier.
Tutorial Dataset¶
For this tutorial, we will be using a subset of the fMRI dataset (ds000114) publicly available on openfmri.org. If you're using the suggested Docker image you probably have all data needed to run the tutorial within the Docker container. If you want to have data locally you can use Datalad to download a subset of the dataset, via the datalad repository. In order to install dataset with all subrepositories you can run:
%%bash
cd /data
datalad install -r ///workshops/nih-2017/ds000114
In order to download data, you can use datalad get foldername command, to download all files in the folder foldername. For this tutorial we only want to download part of the dataset, i.e. the anatomical and the functional fingerfootlips images:
%%bash
cd /data/ds000114
datalad get -J 4 /data/ds000114/derivatives/fmriprep/sub-*/anat/*preproc.nii.gz \
/data/ds000114/sub-01/ses-test/anat \
/data/ds000114/sub-*/ses-test/func/*fingerfootlips*
So let's have a look at the tutorial dataset.
ls /data/ds000114
ls /data/ds000114/sub-01/ses-test/*
As you can, for every subject we have one anatomical T1w image, five functional images, and one diffusion weighted image.
Note: If you used datalad or git annex to get the dataset, you can see symlinks for the image files.
Behavioral Task¶
Subject from the ds000114 dataset did five behavioral tasks. In our dataset two of them are included.
The motor task consisted of finger tapping, foot twitching and lip pouching interleaved with fixation at a cross.
The landmark task was designed to mimic the line bisection task used in neurological practice to diagnose spatial hemineglect. Two conditions were contrasted, specifically judging if a horizontal line had been bisected exactly in the middle, versus judging if a horizontal line was bisected at all. More about the dataset and studies you can find here.
To each of the functional images above, we therefore also have a tab-separated values file (tva), containing information such as stimuli onset, duration, type, etc. So let's have a look at one of them:
%%bash
cd /data/ds000114
datalad get /data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-linebisection_events.tsv
!cat /data/ds000114/sub-01/ses-test/func/sub-01_ses-test_task-linebisection_events.tsv